How Well do Land Classification Algorithms Detect Community Structure Patterns in Forest Ecosystems?
Last fall I had a meeting with Jean Burns and her former undergraduate student, India Johnson, about using remote sensing to study invasive species. Among various options, we discussed using artificial intelligence methods to identify and demarcate abundance and distribution patterns of invasive species. I had already had some experience with semi-automatic classification of land cover using the SCP Plug-in module for QGIS (Congedo, Luca, (2021). Semi-Automatic Classification Plugin: A Python tool for the download and processing of remote sensing images in QGIS. Journal of Open Source Software, 6(64), 3172, https://doi.org/10.21105/joss.03172), and I hadn't had much success with identifying patterns of distribution of the dominant tree species in the north woodlot of CWRU's University Farms. With this post, I will provide an initial assessment of the limitations of land classification methods when applied to forested ecosystems.
For this assessment, I am using two different sources of multispectral imagery (Figs. 1 and 2). Most of the UAV images are from a Sentera Double 4K Multispectral Sensor with RGB (narrow band red, green, and blue bands) and NIR (red edge and near infrared bands) cameras. At 150 m, these images have a pixel resolution of 3 cm and the raster has about 240 million pixels. I have recently discovered the availability of high resolution, multispectral images from Planet (Planet Application Program Interface: In Space for Life on Earth. San Francisco, CA. https://api.planet.com). The image in Fig. 2 contains about 500,000 pixels with a resolution of 3m. There are obvious patterns of land cover in both images, and there appear to be discernible patterns of distribution of different tree species in the high resolution image of the north woodlot (Fig. 1).
 |
Figure 2. Satellite image of the University Farm property (outlined in red). Satellite image is a PSScene4Band image from Planet on June 8, 2020, with a pixel size of 3 m. |
The SCP module in QGIS has two methods of generating land cover classifications. Supervised classifications require an a priori training input to establish spectral signatures of land cover categories. In contrast, unsupervised classifications builds a land cover classification by clustering pixels by similarity of spectral signatures. Fig. 3 is an example of a supervised classification of the north woodlot. Although the patterns are similar, the classification produces a different distribution of forest features. The main obstacle to improving the classification is substantial overlap of spectral signatures of the various tree species used in the training input.
Fig. 4 provides an example of unsupervised classification of the image in Figure 1. A challenge with is identifying land cover categories associated with clusters arising from the unsupervised classification. Areas of grass cover or roads and other built features are easy to identify, but the structure of the forest canopy is much harder to classify by species. Gaps in the forest canopy, however, are well demarcated. High resolution of the image, in fact, leads to detection of gaps in the upper canopy of individual trees.
 |
| Figure 4. Results of supervised classification of forest features from image in Fig. 1. |
The 3 m pixel resolution of the Planet satellite imagers provides a lower resolution option for exploring forest structure. The feature categories in Fig. 5 lack correspondence with features in Fig. 4, except for gap areas. The lower resolution satellite images result in identification of the larger areas between individual tree canopies. The also reveal areas of similar spectral signatures that may not be species specific.
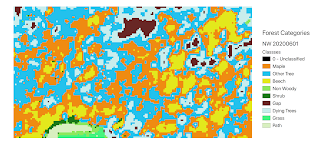 |
| Figure 5. Results of unsupervised classification of forest features in the north woodlot from PSScene4Band image in Fig. 2. |
The reason for the lack of species specific spectral signatures in either Fig. 4 or Fig. 5 likely involves variation in the observable structure of tree canopies. Fig 6, is a high resolution image of 76 x 57 m area within the north woodlot. At this scale of resolution, it is possible to identify maple and oak leaves. Although there is a slight difference in the perceived color of maple and oak branches, the coloration is not constant at different heights in the overall canopy. Thus spectral signatures of lower leaves in the canopy will be different from upper leaves. Also, the gaps between branches is not species specific. Both supervised and unsupervised classifications are limited by the overlap of spectral signatures associated with the physical structure of individual tree canopies.
Conclusion
Although individual leaves of tree species in the north woodlot are easy to distinguish by size, shape, and color, demarcation of individual tree species with remote sensing methods seems impractical. The substantial overlap of spectral signatures of different species leads to unreliable categorization. Because much of the overlap arises from the physical structure of the canopy, however, some categories will be more consistently identified. For example, gap structure of the canopy varies with resolution of images, but is largely consistent (compare Figs. 4 and 5 for location of major canopy gaps). Uppermost leaves of some tree species likely have distinctive spectral signatures. Determining whether those are sufficiently distinctive of account for the other patterns in Figs 4 and 5 remains a work in progress.


